| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-83 | -1.914e+02 | 0.0000 | 828.0 | 27.60% | 220.0 | 8.22% | motif file (matrix) |
pdf |
| 2 |  | GATA(Zf),IR3/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-70 | -1.631e+02 | 0.0000 | 326.0 | 10.87% | 17.7 | 0.66% | motif file (matrix) |
pdf |
| 3 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-66 | -1.536e+02 | 0.0000 | 633.0 | 21.10% | 153.3 | 5.73% | motif file (matrix) |
pdf |
| 4 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-63 | -1.464e+02 | 0.0000 | 546.0 | 18.20% | 116.4 | 4.35% | motif file (matrix) |
pdf |
| 5 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-61 | -1.422e+02 | 0.0000 | 554.0 | 18.47% | 124.0 | 4.63% | motif file (matrix) |
pdf |
| 6 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-59 | -1.373e+02 | 0.0000 | 1046.0 | 34.87% | 429.6 | 16.05% | motif file (matrix) |
pdf |
| 7 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-56 | -1.302e+02 | 0.0000 | 597.0 | 19.90% | 159.9 | 5.97% | motif file (matrix) |
pdf |
| 8 |  | GATA(Zf),IR4/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-55 | -1.279e+02 | 0.0000 | 256.0 | 8.53% | 13.9 | 0.52% | motif file (matrix) |
pdf |
| 9 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-52 | -1.203e+02 | 0.0000 | 450.0 | 15.00% | 94.9 | 3.54% | motif file (matrix) |
pdf |
| 10 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-49 | -1.135e+02 | 0.0000 | 407.0 | 13.57% | 80.4 | 3.00% | motif file (matrix) |
pdf |
| 11 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-45 | -1.052e+02 | 0.0000 | 440.0 | 14.67% | 104.2 | 3.89% | motif file (matrix) |
pdf |
| 12 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-43 | -1.002e+02 | 0.0000 | 370.0 | 12.33% | 75.1 | 2.81% | motif file (matrix) |
pdf |
| 13 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-33 | -7.635e+01 | 0.0000 | 349.0 | 11.63% | 89.7 | 3.35% | motif file (matrix) |
pdf |
| 14 |  | GATA3(Zf),DR8/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-31 | -7.226e+01 | 0.0000 | 155.0 | 5.17% | 10.7 | 0.40% | motif file (matrix) |
pdf |
| 15 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-28 | -6.489e+01 | 0.0000 | 268.0 | 8.93% | 61.1 | 2.28% | motif file (matrix) |
pdf |
| 16 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-27 | -6.433e+01 | 0.0000 | 496.0 | 16.53% | 190.9 | 7.13% | motif file (matrix) |
pdf |
| 17 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-26 | -6.064e+01 | 0.0000 | 225.0 | 7.50% | 45.5 | 1.70% | motif file (matrix) |
pdf |
| 18 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-25 | -5.980e+01 | 0.0000 | 203.0 | 6.77% | 36.7 | 1.37% | motif file (matrix) |
pdf |
| 19 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-25 | -5.887e+01 | 0.0000 | 257.0 | 8.57% | 62.9 | 2.35% | motif file (matrix) |
pdf |
| 20 |  | TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer | 1e-24 | -5.697e+01 | 0.0000 | 176.0 | 5.87% | 27.9 | 1.04% | motif file (matrix) |
pdf |
| 21 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-24 | -5.599e+01 | 0.0000 | 211.0 | 7.03% | 43.3 | 1.62% | motif file (matrix) |
pdf |
| 22 |  | PQM-1(?)/cElegans-L3-ChIP-Seq(modEncode)/Homer | 1e-18 | -4.262e+01 | 0.0000 | 174.0 | 5.80% | 39.5 | 1.47% | motif file (matrix) |
pdf |
| 23 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-17 | -3.980e+01 | 0.0000 | 188.0 | 6.27% | 49.4 | 1.85% | motif file (matrix) |
pdf |
| 24 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer | 1e-17 | -3.958e+01 | 0.0000 | 67.0 | 2.23% | 1.7 | 0.06% | motif file (matrix) |
pdf |
| 25 |  | ELT-3(Gata)/cElegans-L1-ELT3-ChIP-Seq(modEncode)/Homer | 1e-14 | -3.231e+01 | 0.0000 | 185.0 | 6.17% | 57.5 | 2.15% | motif file (matrix) |
pdf |
| 26 |  | GATA3(Zf),DR4/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-13 | -3.144e+01 | 0.0000 | 104.0 | 3.47% | 18.7 | 0.70% | motif file (matrix) |
pdf |
| 27 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-11 | -2.567e+01 | 0.0000 | 146.0 | 4.87% | 45.9 | 1.72% | motif file (matrix) |
pdf |
| 28 |  | Unknown5/Drosophila-Promoters/Homer | 1e-11 | -2.551e+01 | 0.0000 | 174.0 | 5.80% | 61.8 | 2.31% | motif file (matrix) |
pdf |
| 29 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-10 | -2.519e+01 | 0.0000 | 64.0 | 2.13% | 7.9 | 0.29% | motif file (matrix) |
pdf |
| 30 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-10 | -2.480e+01 | 0.0000 | 212.0 | 7.07% | 85.7 | 3.20% | motif file (matrix) |
pdf |
| 31 | 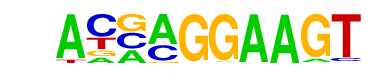 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-10 | -2.321e+01 | 0.0000 | 152.0 | 5.07% | 52.0 | 1.94% | motif file (matrix) |
pdf |
| 32 | 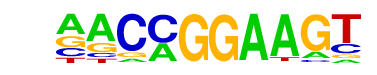 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-9 | -2.236e+01 | 0.0000 | 217.0 | 7.23% | 93.6 | 3.50% | motif file (matrix) |
pdf |
| 33 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-9 | -2.192e+01 | 0.0000 | 157.0 | 5.23% | 57.7 | 2.16% | motif file (matrix) |
pdf |
| 34 | 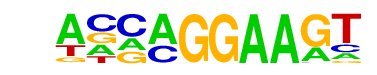 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-9 | -2.084e+01 | 0.0000 | 235.0 | 7.83% | 108.9 | 4.07% | motif file (matrix) |
pdf |
| 35 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-8 | -1.931e+01 | 0.0000 | 237.0 | 7.90% | 113.2 | 4.23% | motif file (matrix) |
pdf |
| 36 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-8 | -1.873e+01 | 0.0000 | 352.0 | 11.73% | 195.7 | 7.31% | motif file (matrix) |
pdf |
| 37 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.662e+01 | 0.0000 | 95.0 | 3.17% | 30.5 | 1.14% | motif file (matrix) |
pdf |
| 38 |  | NF1(CTF)/LNCAP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-7 | -1.661e+01 | 0.0000 | 80.0 | 2.67% | 22.5 | 0.84% | motif file (matrix) |
pdf |
| 39 | 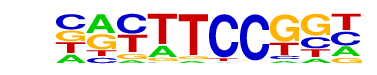 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-7 | -1.660e+01 | 0.0000 | 168.0 | 5.60% | 74.1 | 2.77% | motif file (matrix) |
pdf |
| 40 |  | Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-7 | -1.614e+01 | 0.0000 | 71.0 | 2.37% | 18.4 | 0.69% | motif file (matrix) |
pdf |
| 41 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-6 | -1.533e+01 | 0.0000 | 73.0 | 2.43% | 20.4 | 0.76% | motif file (matrix) |
pdf |
| 42 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-5 | -1.259e+01 | 0.0000 | 68.0 | 2.27% | 21.2 | 0.79% | motif file (matrix) |
pdf |
| 43 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-5 | -1.254e+01 | 0.0000 | 210.0 | 7.00% | 113.2 | 4.23% | motif file (matrix) |
pdf |
| 44 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-5 | -1.203e+01 | 0.0001 | 61.0 | 2.03% | 18.4 | 0.69% | motif file (matrix) |
pdf |
| 45 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-5 | -1.190e+01 | 0.0001 | 136.0 | 4.53% | 64.1 | 2.40% | motif file (matrix) |
pdf |
| 46 |  | SeqBias: polyA-repeat | 1e-4 | -1.123e+01 | 0.0001 | 2917.0 | 97.23% | 2544.3 | 95.05% | motif file (matrix) |
pdf |
| 47 |  | GATA:SCL(Zf,bHLH)/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-4 | -1.102e+01 | 0.0001 | 45.0 | 1.50% | 11.7 | 0.44% | motif file (matrix) |
pdf |
| 48 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.098e+01 | 0.0001 | 69.0 | 2.30% | 24.5 | 0.92% | motif file (matrix) |
pdf |
| 49 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-4 | -1.057e+01 | 0.0002 | 122.0 | 4.07% | 58.4 | 2.18% | motif file (matrix) |
pdf |
| 50 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-4 | -1.016e+01 | 0.0003 | 43.0 | 1.43% | 12.0 | 0.45% | motif file (matrix) |
pdf |
| 51 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.961e+00 | 0.0004 | 52.0 | 1.73% | 16.6 | 0.62% | motif file (matrix) |
pdf |
| 52 |  | ETS(ETS)/Promoter/Homer | 1e-4 | -9.578e+00 | 0.0005 | 51.0 | 1.70% | 16.4 | 0.61% | motif file (matrix) |
pdf |
| 53 |  | HOXB13(Homeobox)/ProstateTumor-HOXB13-ChIP-Seq(GSE56288)/Homer | 1e-4 | -9.535e+00 | 0.0005 | 234.0 | 7.80% | 141.7 | 5.29% | motif file (matrix) |
pdf |
| 54 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-4 | -9.507e+00 | 0.0005 | 109.0 | 3.63% | 52.0 | 1.94% | motif file (matrix) |
pdf |
| 55 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.479e+00 | 0.0005 | 63.0 | 2.10% | 23.9 | 0.89% | motif file (matrix) |
pdf |
| 56 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.224e+00 | 0.0007 | 130.0 | 4.33% | 67.9 | 2.54% | motif file (matrix) |
pdf |
| 57 |  | SeqBias: A/T bias | 1e-3 | -8.773e+00 | 0.0011 | 2393.0 | 79.77% | 2027.7 | 75.75% | motif file (matrix) |
pdf |
| 58 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-3 | -8.455e+00 | 0.0014 | 102.0 | 3.40% | 50.7 | 1.89% | motif file (matrix) |
pdf |
| 59 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-3 | -8.323e+00 | 0.0016 | 747.0 | 24.90% | 561.9 | 20.99% | motif file (matrix) |
pdf |
| 60 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.137e+00 | 0.0019 | 59.0 | 1.97% | 23.4 | 0.87% | motif file (matrix) |
pdf |
| 61 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.095e+00 | 0.0019 | 105.0 | 3.50% | 53.8 | 2.01% | motif file (matrix) |
pdf |
| 62 |  | NF1:FOXA1(CTF,Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-3 | -8.040e+00 | 0.0020 | 28.0 | 0.93% | 6.0 | 0.23% | motif file (matrix) |
pdf |
| 63 |  | Tal1 | 1e-3 | -7.845e+00 | 0.0024 | 301.0 | 10.03% | 200.9 | 7.51% | motif file (matrix) |
pdf |
| 64 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.427e+00 | 0.0036 | 71.0 | 2.37% | 32.8 | 1.22% | motif file (matrix) |
pdf |
| 65 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-3 | -7.422e+00 | 0.0036 | 108.0 | 3.60% | 57.9 | 2.16% | motif file (matrix) |
pdf |
| 66 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-3 | -7.001e+00 | 0.0053 | 37.0 | 1.23% | 12.7 | 0.47% | motif file (matrix) |
pdf |
| 67 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-2 | -6.856e+00 | 0.0061 | 114.0 | 3.80% | 63.9 | 2.39% | motif file (matrix) |
pdf |
| 68 |  | Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.618e+00 | 0.0076 | 62.0 | 2.07% | 28.7 | 1.07% | motif file (matrix) |
pdf |
| 69 |  | Zelda(Zf)/Embryo-zld-ChIP-Seq(GSE65441)/Homer | 1e-2 | -6.319e+00 | 0.0101 | 67.0 | 2.23% | 32.4 | 1.21% | motif file (matrix) |
pdf |
| 70 |  | Atf7(bZIP)/3T3L1-Atf7-ChIP-Seq(GSE56872)/Homer | 1e-2 | -5.845e+00 | 0.0160 | 74.0 | 2.47% | 38.8 | 1.45% | motif file (matrix) |
pdf |
| 71 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-2 | -5.816e+00 | 0.0162 | 98.0 | 3.27% | 55.6 | 2.08% | motif file (matrix) |
pdf |
| 72 |  | ERE(NR),IR3/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.799e+00 | 0.0163 | 32.0 | 1.07% | 11.5 | 0.43% | motif file (matrix) |
pdf |
| 73 |  | PR(NR)/T47D-PR-ChIP-Seq(GSE31130)/Homer | 1e-2 | -5.717e+00 | 0.0174 | 287.0 | 9.57% | 201.6 | 7.53% | motif file (matrix) |
pdf |
| 74 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-2 | -5.490e+00 | 0.0216 | 162.0 | 5.40% | 104.2 | 3.89% | motif file (matrix) |
pdf |
| 75 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -5.199e+00 | 0.0285 | 11.0 | 0.37% | 2.0 | 0.07% | motif file (matrix) |
pdf |
| 76 |  | Atf2(bZIP)/3T3L1-Atf2-ChIP-Seq(GSE56872)/Homer | 1e-2 | -5.106e+00 | 0.0309 | 49.0 | 1.63% | 23.4 | 0.87% | motif file (matrix) |
pdf |
| 77 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-2 | -5.105e+00 | 0.0309 | 8.0 | 0.27% | 0.0 | 0.00% | motif file (matrix) |
pdf |

























































































































































