| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Mef2b(MADS)/HEK293-Mef2b.V5-ChIP-Seq(GSE67450)/Homer | 1e-299 | -6.893e+02 | 0.0000 | 1403.0 | 46.77% | 153.6 | 5.64% | motif file (matrix) |
pdf |
| 2 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-225 | -5.188e+02 | 0.0000 | 1090.0 | 36.33% | 108.8 | 4.00% | motif file (matrix) |
pdf |
| 3 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529)/Homer | 1e-189 | -4.362e+02 | 0.0000 | 960.0 | 32.00% | 100.2 | 3.68% | motif file (matrix) |
pdf |
| 4 |  | Mef2d(MADS)/Retina-Mef2d-ChIP-Seq(GSE61391)/Homer | 1e-169 | -3.910e+02 | 0.0000 | 719.0 | 23.97% | 36.1 | 1.33% | motif file (matrix) |
pdf |
| 5 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-77 | -1.788e+02 | 0.0000 | 407.0 | 13.57% | 36.8 | 1.35% | motif file (matrix) |
pdf |
| 6 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-75 | -1.746e+02 | 0.0000 | 420.0 | 14.00% | 43.4 | 1.59% | motif file (matrix) |
pdf |
| 7 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-72 | -1.661e+02 | 0.0000 | 369.0 | 12.30% | 30.0 | 1.10% | motif file (matrix) |
pdf |
| 8 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-67 | -1.552e+02 | 0.0000 | 404.0 | 13.47% | 49.5 | 1.82% | motif file (matrix) |
pdf |
| 9 |  | SOC1(MADS)/Seedling-SOC1-ChIP-Seq(GSE45846)/Homer | 1e-52 | -1.199e+02 | 0.0000 | 315.0 | 10.50% | 38.6 | 1.42% | motif file (matrix) |
pdf |
| 10 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-46 | -1.065e+02 | 0.0000 | 229.0 | 7.63% | 16.2 | 0.60% | motif file (matrix) |
pdf |
| 11 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-44 | -1.021e+02 | 0.0000 | 394.0 | 13.13% | 87.1 | 3.20% | motif file (matrix) |
pdf |
| 12 |  | SEP3(MADS)/Arabidoposis-Flower-Sep3-ChIP-Seq/Homer | 1e-43 | -1.008e+02 | 0.0000 | 634.0 | 21.13% | 223.5 | 8.21% | motif file (matrix) |
pdf |
| 13 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-40 | -9.291e+01 | 0.0000 | 183.0 | 6.10% | 9.5 | 0.35% | motif file (matrix) |
pdf |
| 14 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-40 | -9.224e+01 | 0.0000 | 313.0 | 10.43% | 58.7 | 2.16% | motif file (matrix) |
pdf |
| 15 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-36 | -8.418e+01 | 0.0000 | 327.0 | 10.90% | 72.7 | 2.67% | motif file (matrix) |
pdf |
| 16 | 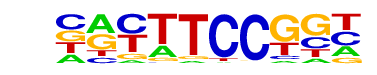 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-31 | -7.264e+01 | 0.0000 | 295.0 | 9.83% | 68.7 | 2.52% | motif file (matrix) |
pdf |
| 17 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-29 | -6.842e+01 | 0.0000 | 278.0 | 9.27% | 64.2 | 2.36% | motif file (matrix) |
pdf |
| 18 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-29 | -6.797e+01 | 0.0000 | 368.0 | 12.27% | 112.8 | 4.14% | motif file (matrix) |
pdf |
| 19 |  | PU.1:IRF8(ETS:IRF)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer | 1e-29 | -6.682e+01 | 0.0000 | 171.0 | 5.70% | 19.7 | 0.72% | motif file (matrix) |
pdf |
| 20 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-28 | -6.654e+01 | 0.0000 | 272.0 | 9.07% | 63.4 | 2.33% | motif file (matrix) |
pdf |
| 21 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-28 | -6.614e+01 | 0.0000 | 244.0 | 8.13% | 50.2 | 1.84% | motif file (matrix) |
pdf |
| 22 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-26 | -6.143e+01 | 0.0000 | 340.0 | 11.33% | 105.1 | 3.86% | motif file (matrix) |
pdf |
| 23 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-25 | -5.795e+01 | 0.0000 | 178.0 | 5.93% | 28.7 | 1.06% | motif file (matrix) |
pdf |
| 24 |  | bZIP:IRF(bZIP,IRF)/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-24 | -5.544e+01 | 0.0000 | 228.0 | 7.60% | 53.5 | 1.97% | motif file (matrix) |
pdf |
| 25 | 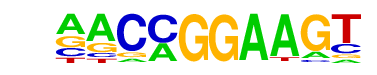 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-23 | -5.437e+01 | 0.0000 | 314.0 | 10.47% | 100.8 | 3.70% | motif file (matrix) |
pdf |
| 26 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-23 | -5.404e+01 | 0.0000 | 225.0 | 7.50% | 54.0 | 1.98% | motif file (matrix) |
pdf |
| 27 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-23 | -5.354e+01 | 0.0000 | 97.0 | 3.23% | 3.3 | 0.12% | motif file (matrix) |
pdf |
| 28 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-21 | -4.855e+01 | 0.0000 | 215.0 | 7.17% | 54.7 | 2.01% | motif file (matrix) |
pdf |
| 29 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-18 | -4.326e+01 | 0.0000 | 128.0 | 4.27% | 19.2 | 0.71% | motif file (matrix) |
pdf |
| 30 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-18 | -4.304e+01 | 0.0000 | 112.0 | 3.73% | 13.3 | 0.49% | motif file (matrix) |
pdf |
| 31 | 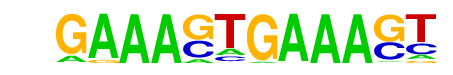 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-17 | -4.062e+01 | 0.0000 | 118.0 | 3.93% | 17.5 | 0.64% | motif file (matrix) |
pdf |
| 32 |  | IRF:BATF(IRF:bZIP)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer | 1e-17 | -4.021e+01 | 0.0000 | 127.0 | 4.23% | 21.2 | 0.78% | motif file (matrix) |
pdf |
| 33 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan_et_al.)/Homer | 1e-16 | -3.890e+01 | 0.0000 | 149.0 | 4.97% | 32.9 | 1.21% | motif file (matrix) |
pdf |
| 34 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-16 | -3.831e+01 | 0.0000 | 182.0 | 6.07% | 49.5 | 1.82% | motif file (matrix) |
pdf |
| 35 | 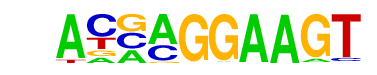 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-16 | -3.741e+01 | 0.0000 | 178.0 | 5.93% | 48.2 | 1.77% | motif file (matrix) |
pdf |
| 36 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-15 | -3.612e+01 | 0.0000 | 94.0 | 3.13% | 11.2 | 0.41% | motif file (matrix) |
pdf |
| 37 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-14 | -3.305e+01 | 0.0000 | 115.0 | 3.83% | 22.1 | 0.81% | motif file (matrix) |
pdf |
| 38 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-14 | -3.293e+01 | 0.0000 | 132.0 | 4.40% | 30.7 | 1.13% | motif file (matrix) |
pdf |
| 39 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-14 | -3.247e+01 | 0.0000 | 131.0 | 4.37% | 30.6 | 1.12% | motif file (matrix) |
pdf |
| 40 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-13 | -3.195e+01 | 0.0000 | 136.0 | 4.53% | 33.7 | 1.24% | motif file (matrix) |
pdf |
| 41 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-13 | -3.150e+01 | 0.0000 | 137.0 | 4.57% | 34.5 | 1.27% | motif file (matrix) |
pdf |
| 42 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-13 | -3.117e+01 | 0.0000 | 244.0 | 8.13% | 95.0 | 3.49% | motif file (matrix) |
pdf |
| 43 |  | ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer | 1e-12 | -2.852e+01 | 0.0000 | 60.0 | 2.00% | 4.9 | 0.18% | motif file (matrix) |
pdf |
| 44 |  | ETS(ETS)/Promoter/Homer | 1e-12 | -2.813e+01 | 0.0000 | 89.0 | 2.97% | 15.6 | 0.57% | motif file (matrix) |
pdf |
| 45 |  | SeqBias: A/T bias | 1e-11 | -2.584e+01 | 0.0000 | 2604.0 | 86.80% | 2181.8 | 80.14% | motif file (matrix) |
pdf |
| 46 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-11 | -2.558e+01 | 0.0000 | 129.0 | 4.30% | 37.5 | 1.38% | motif file (matrix) |
pdf |
| 47 |  | ETS:RUNX(ETS,Runt)/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-10 | -2.426e+01 | 0.0000 | 46.0 | 1.53% | 2.8 | 0.10% | motif file (matrix) |
pdf |
| 48 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.269e+01 | 0.0000 | 35.0 | 1.17% | 0.5 | 0.02% | motif file (matrix) |
pdf |
| 49 |  | PRDM1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.235e+01 | 0.0000 | 161.0 | 5.37% | 60.6 | 2.23% | motif file (matrix) |
pdf |
| 50 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-9 | -2.139e+01 | 0.0000 | 33.0 | 1.10% | 0.5 | 0.02% | motif file (matrix) |
pdf |
| 51 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.692e+01 | 0.0000 | 37.0 | 1.23% | 3.0 | 0.11% | motif file (matrix) |
pdf |
| 52 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.671e+01 | 0.0000 | 30.0 | 1.00% | 1.9 | 0.07% | motif file (matrix) |
pdf |
| 53 |  | SeqBias: polyA-repeat | 1e-7 | -1.636e+01 | 0.0000 | 2908.0 | 96.93% | 2560.8 | 94.06% | motif file (matrix) |
pdf |
| 54 |  | Initiator/Drosophila-Promoters/Homer | 1e-6 | -1.610e+01 | 0.0000 | 481.0 | 16.03% | 307.3 | 11.29% | motif file (matrix) |
pdf |
| 55 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-6 | -1.591e+01 | 0.0000 | 104.0 | 3.47% | 37.0 | 1.36% | motif file (matrix) |
pdf |
| 56 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.587e+01 | 0.0000 | 143.0 | 4.77% | 61.3 | 2.25% | motif file (matrix) |
pdf |
| 57 |  | Tcf12(bHLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-6 | -1.543e+01 | 0.0000 | 140.0 | 4.67% | 60.7 | 2.23% | motif file (matrix) |
pdf |
| 58 |  | MyoG(bHLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-6 | -1.531e+01 | 0.0000 | 152.0 | 5.07% | 68.7 | 2.52% | motif file (matrix) |
pdf |
| 59 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-6 | -1.440e+01 | 0.0000 | 108.0 | 3.60% | 42.7 | 1.57% | motif file (matrix) |
pdf |
| 60 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-5 | -1.355e+01 | 0.0000 | 50.0 | 1.67% | 11.1 | 0.41% | motif file (matrix) |
pdf |
| 61 |  | Ap4(bHLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-5 | -1.339e+01 | 0.0000 | 169.0 | 5.63% | 84.9 | 3.12% | motif file (matrix) |
pdf |
| 62 |  | Atf2(bZIP)/3T3L1-Atf2-ChIP-Seq(GSE56872)/Homer | 1e-5 | -1.314e+01 | 0.0000 | 61.0 | 2.03% | 17.4 | 0.64% | motif file (matrix) |
pdf |
| 63 |  | Tcf21(bHLH)/ArterySmoothMuscle-Tcf21-ChIP-Seq(GSE61369)/Homer | 1e-5 | -1.288e+01 | 0.0000 | 136.0 | 4.53% | 63.6 | 2.33% | motif file (matrix) |
pdf |
| 64 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-5 | -1.270e+01 | 0.0000 | 185.0 | 6.17% | 97.4 | 3.58% | motif file (matrix) |
pdf |
| 65 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.243e+01 | 0.0000 | 63.0 | 2.10% | 19.9 | 0.73% | motif file (matrix) |
pdf |
| 66 |  | NFkB-p65-Rel(RHD)/ThioMac-LPS-Expression(GSE23622)/Homer | 1e-5 | -1.179e+01 | 0.0000 | 33.0 | 1.10% | 5.5 | 0.20% | motif file (matrix) |
pdf |
| 67 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-5 | -1.176e+01 | 0.0000 | 44.0 | 1.47% | 10.7 | 0.39% | motif file (matrix) |
pdf |
| 68 |  | Atf7(bZIP)/3T3L1-Atf7-ChIP-Seq(GSE56872)/Homer | 1e-4 | -1.123e+01 | 0.0001 | 95.0 | 3.17% | 40.6 | 1.49% | motif file (matrix) |
pdf |
| 69 |  | Ascl1(bHLH)/NeuralTubes-Ascl1-ChIP-Seq(GSE55840)/Homer | 1e-4 | -1.120e+01 | 0.0001 | 194.0 | 6.47% | 108.8 | 3.99% | motif file (matrix) |
pdf |
| 70 |  | Oct6(POU,Homeobox)/NPC-Oct6-ChIP-Seq(GSE35496)/Homer | 1e-4 | -1.015e+01 | 0.0002 | 74.0 | 2.47% | 29.6 | 1.09% | motif file (matrix) |
pdf |
| 71 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-4 | -9.949e+00 | 0.0003 | 281.0 | 9.37% | 178.9 | 6.57% | motif file (matrix) |
pdf |
| 72 |  | Ptf1a(bHLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-4 | -9.620e+00 | 0.0004 | 348.0 | 11.60% | 232.5 | 8.54% | motif file (matrix) |
pdf |
| 73 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.317e+00 | 0.0005 | 127.0 | 4.23% | 66.2 | 2.43% | motif file (matrix) |
pdf |
| 74 |  | Sp1(Zf)/Promoter/Homer | 1e-3 | -8.877e+00 | 0.0007 | 20.0 | 0.67% | 2.9 | 0.11% | motif file (matrix) |
pdf |
| 75 |  | Dorsal(RHD)/Embryo-dl-ChIP-Seq(GSE65441)/Homer | 1e-3 | -8.782e+00 | 0.0008 | 41.0 | 1.37% | 12.3 | 0.45% | motif file (matrix) |
pdf |
| 76 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.534e+00 | 0.0010 | 128.0 | 4.27% | 69.1 | 2.54% | motif file (matrix) |
pdf |
| 77 |  | E2A(bHLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-3 | -7.966e+00 | 0.0017 | 197.0 | 6.57% | 122.6 | 4.50% | motif file (matrix) |
pdf |
| 78 |  | Unknown1/Arabidopsis-Promoters/Homer | 1e-3 | -7.796e+00 | 0.0020 | 25.0 | 0.83% | 5.8 | 0.21% | motif file (matrix) |
pdf |
| 79 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-3 | -7.759e+00 | 0.0021 | 12.0 | 0.40% | 0.7 | 0.03% | motif file (matrix) |
pdf |
| 80 |  | HLH-1(bHLH)/cElegans-Embryo-HLH1-ChIP-Seq(modEncode)/Homer | 1e-3 | -7.572e+00 | 0.0025 | 107.0 | 3.57% | 57.2 | 2.10% | motif file (matrix) |
pdf |
| 81 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-3 | -7.525e+00 | 0.0026 | 20.0 | 0.67% | 3.4 | 0.12% | motif file (matrix) |
pdf |
| 82 |  | Brn1(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer | 1e-3 | -7.306e+00 | 0.0032 | 54.0 | 1.80% | 22.2 | 0.82% | motif file (matrix) |
pdf |
| 83 |  | NFkB-p50,p52(RHD)/Monocyte-p50-ChIP-Chip(Schreiber_et_al.)/Homer | 1e-3 | -7.295e+00 | 0.0032 | 26.0 | 0.87% | 6.1 | 0.23% | motif file (matrix) |
pdf |
| 84 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-3 | -7.243e+00 | 0.0033 | 296.0 | 9.87% | 203.3 | 7.47% | motif file (matrix) |
pdf |
| 85 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-3 | -7.160e+00 | 0.0035 | 575.0 | 19.17% | 434.9 | 15.97% | motif file (matrix) |
pdf |
| 86 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-3 | -6.922e+00 | 0.0044 | 48.0 | 1.60% | 19.5 | 0.72% | motif file (matrix) |
pdf |
| 87 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -6.899e+00 | 0.0045 | 21.0 | 0.70% | 4.1 | 0.15% | motif file (matrix) |
pdf |
| 88 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -6.778e+00 | 0.0050 | 41.0 | 1.37% | 15.0 | 0.55% | motif file (matrix) |
pdf |
| 89 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.585e+00 | 0.0060 | 126.0 | 4.20% | 74.9 | 2.75% | motif file (matrix) |
pdf |
| 90 |  | PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer | 1e-2 | -6.454e+00 | 0.0068 | 59.0 | 1.97% | 27.6 | 1.01% | motif file (matrix) |
pdf |
| 91 |  | PAX6(Paired,Homeobox)/Forebrain-Pax6-ChIP-Seq(GSE66961)/Homer | 1e-2 | -6.134e+00 | 0.0092 | 15.0 | 0.50% | 2.4 | 0.09% | motif file (matrix) |
pdf |
| 92 |  | Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.049e+00 | 0.0099 | 65.0 | 2.17% | 32.2 | 1.18% | motif file (matrix) |
pdf |
| 93 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-2 | -5.495e+00 | 0.0171 | 95.0 | 3.17% | 55.5 | 2.04% | motif file (matrix) |
pdf |
| 94 |  | CHR(?)/Hela-CellCycle-Expression/Homer | 1e-2 | -5.466e+00 | 0.0174 | 107.0 | 3.57% | 64.8 | 2.38% | motif file (matrix) |
pdf |
| 95 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-2 | -5.134e+00 | 0.0240 | 156.0 | 5.20% | 103.3 | 3.80% | motif file (matrix) |
pdf |
| 96 |  | SeqBias: TA-repeat | 1e-2 | -5.094e+00 | 0.0247 | 885.0 | 29.50% | 721.9 | 26.52% | motif file (matrix) |
pdf |
| 97 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.059e+00 | 0.0254 | 64.0 | 2.13% | 35.0 | 1.28% | motif file (matrix) |
pdf |
| 98 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.048e+00 | 0.0254 | 39.0 | 1.30% | 18.0 | 0.66% | motif file (matrix) |
pdf |
| 99 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-2 | -5.019e+00 | 0.0258 | 127.0 | 4.23% | 81.5 | 2.99% | motif file (matrix) |
pdf |
| 100 |  | HEB(bHLH)/mES-Heb-ChIP-Seq(GSE53233)/Homer | 1e-2 | -5.011e+00 | 0.0258 | 234.0 | 7.80% | 166.8 | 6.13% | motif file (matrix) |
pdf |
| 101 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-2 | -4.961e+00 | 0.0268 | 111.0 | 3.70% | 69.2 | 2.54% | motif file (matrix) |
pdf |
| 102 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -4.799e+00 | 0.0313 | 269.0 | 8.97% | 196.4 | 7.21% | motif file (matrix) |
pdf |
| 103 |  | NRF(NRF)/Promoter/Homer | 1e-2 | -4.712e+00 | 0.0338 | 10.0 | 0.33% | 1.5 | 0.05% | motif file (matrix) |
pdf |
| 104 |  | PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-2 | -4.698e+00 | 0.0339 | 79.0 | 2.63% | 46.7 | 1.71% | motif file (matrix) |
pdf |
| 105 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-2 | -4.695e+00 | 0.0339 | 20.0 | 0.67% | 7.0 | 0.26% | motif file (matrix) |
pdf |

















































































































































































































