| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-88 | -2.045e+02 | 0.0000 | 765.0 | 25.50% | 126.8 | 5.65% | motif file (matrix) |
pdf |
| 2 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-72 | -1.662e+02 | 0.0000 | 655.0 | 21.83% | 112.3 | 5.00% | motif file (matrix) |
pdf |
| 3 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-63 | -1.464e+02 | 0.0000 | 621.0 | 20.70% | 116.0 | 5.17% | motif file (matrix) |
pdf |
| 4 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-60 | -1.399e+02 | 0.0000 | 448.0 | 14.93% | 53.7 | 2.39% | motif file (matrix) |
pdf |
| 5 |  | Zic(Zf)/Cerebellum-ZIC1.2-ChIP-Seq(GSE60731)/Homer | 1e-57 | -1.326e+02 | 0.0000 | 456.0 | 15.20% | 61.3 | 2.73% | motif file (matrix) |
pdf |
| 6 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-56 | -1.306e+02 | 0.0000 | 388.0 | 12.93% | 39.5 | 1.76% | motif file (matrix) |
pdf |
| 7 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-51 | -1.176e+02 | 0.0000 | 1151.0 | 38.37% | 433.0 | 19.28% | motif file (matrix) |
pdf |
| 8 |  | OCT4-SOX2-TCF-NANOG(POU,Homeobox,HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-47 | -1.100e+02 | 0.0000 | 267.0 | 8.90% | 15.6 | 0.70% | motif file (matrix) |
pdf |
| 9 |  | Sox15(HMG)/CPA-Sox15-ChIP-Seq(GSE62909)/Homer | 1e-40 | -9.358e+01 | 0.0000 | 433.0 | 14.43% | 85.0 | 3.79% | motif file (matrix) |
pdf |
| 10 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-34 | -7.871e+01 | 0.0000 | 338.0 | 11.27% | 60.3 | 2.69% | motif file (matrix) |
pdf |
| 11 |  | Sox9(HMG)/Limb-SOX9-ChIP-Seq(GSE73225)/Homer | 1e-33 | -7.697e+01 | 0.0000 | 298.0 | 9.93% | 46.5 | 2.07% | motif file (matrix) |
pdf |
| 12 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-30 | -6.909e+01 | 0.0000 | 572.0 | 19.07% | 182.5 | 8.13% | motif file (matrix) |
pdf |
| 13 |  | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-28 | -6.510e+01 | 0.0000 | 228.0 | 7.60% | 30.2 | 1.35% | motif file (matrix) |
pdf |
| 14 |  | Oct6(POU,Homeobox)/NPC-Oct6-ChIP-Seq(GSE35496)/Homer | 1e-23 | -5.459e+01 | 0.0000 | 188.0 | 6.27% | 24.1 | 1.07% | motif file (matrix) |
pdf |
| 15 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-18 | -4.339e+01 | 0.0000 | 649.0 | 21.63% | 274.4 | 12.22% | motif file (matrix) |
pdf |
| 16 |  | Brn1(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer | 1e-18 | -4.147e+01 | 0.0000 | 142.0 | 4.73% | 18.7 | 0.83% | motif file (matrix) |
pdf |
| 17 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-11 | -2.561e+01 | 0.0000 | 957.0 | 31.90% | 526.5 | 23.45% | motif file (matrix) |
pdf |
| 18 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-10 | -2.438e+01 | 0.0000 | 188.0 | 6.27% | 56.1 | 2.50% | motif file (matrix) |
pdf |
| 19 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-10 | -2.436e+01 | 0.0000 | 413.0 | 13.77% | 180.7 | 8.05% | motif file (matrix) |
pdf |
| 20 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-10 | -2.320e+01 | 0.0000 | 110.0 | 3.67% | 22.5 | 1.00% | motif file (matrix) |
pdf |
| 21 |  | Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-9 | -2.237e+01 | 0.0000 | 103.0 | 3.43% | 20.1 | 0.90% | motif file (matrix) |
pdf |
| 22 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-9 | -2.089e+01 | 0.0000 | 142.0 | 4.73% | 39.5 | 1.76% | motif file (matrix) |
pdf |
| 23 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-9 | -2.073e+01 | 0.0000 | 122.0 | 4.07% | 30.5 | 1.36% | motif file (matrix) |
pdf |
| 24 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-8 | -2.071e+01 | 0.0000 | 275.0 | 9.17% | 109.5 | 4.88% | motif file (matrix) |
pdf |
| 25 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-8 | -1.880e+01 | 0.0000 | 154.0 | 5.13% | 48.3 | 2.15% | motif file (matrix) |
pdf |
| 26 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-8 | -1.846e+01 | 0.0000 | 184.0 | 6.13% | 64.3 | 2.86% | motif file (matrix) |
pdf |
| 27 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-7 | -1.825e+01 | 0.0000 | 136.0 | 4.53% | 40.7 | 1.81% | motif file (matrix) |
pdf |
| 28 |  | TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer | 1e-7 | -1.770e+01 | 0.0000 | 126.0 | 4.20% | 36.6 | 1.63% | motif file (matrix) |
pdf |
| 29 |  | Tcf12(bHLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.728e+01 | 0.0000 | 202.0 | 6.73% | 76.5 | 3.41% | motif file (matrix) |
pdf |
| 30 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer | 1e-7 | -1.629e+01 | 0.0000 | 38.0 | 1.27% | 2.8 | 0.12% | motif file (matrix) |
pdf |
| 31 |  | Lhx1(Homeobox)/EmbryoCarcinoma-Lhx1-ChIP-Seq(GSE70957)/Homer | 1e-7 | -1.620e+01 | 0.0000 | 280.0 | 9.33% | 123.4 | 5.50% | motif file (matrix) |
pdf |
| 32 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-6 | -1.608e+01 | 0.0000 | 139.0 | 4.63% | 45.7 | 2.03% | motif file (matrix) |
pdf |
| 33 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-6 | -1.568e+01 | 0.0000 | 155.0 | 5.17% | 54.6 | 2.43% | motif file (matrix) |
pdf |
| 34 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-6 | -1.526e+01 | 0.0000 | 142.0 | 4.73% | 48.3 | 2.15% | motif file (matrix) |
pdf |
| 35 | 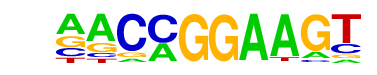 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-6 | -1.461e+01 | 0.0000 | 186.0 | 6.20% | 73.2 | 3.26% | motif file (matrix) |
pdf |
| 36 |  | Ap4(bHLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-5 | -1.378e+01 | 0.0000 | 209.0 | 6.97% | 88.5 | 3.94% | motif file (matrix) |
pdf |
| 37 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-5 | -1.368e+01 | 0.0000 | 121.0 | 4.03% | 40.7 | 1.81% | motif file (matrix) |
pdf |
| 38 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-5 | -1.314e+01 | 0.0000 | 67.0 | 2.23% | 15.7 | 0.70% | motif file (matrix) |
pdf |
| 39 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-5 | -1.297e+01 | 0.0000 | 214.0 | 7.13% | 93.2 | 4.15% | motif file (matrix) |
pdf |
| 40 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-5 | -1.254e+01 | 0.0000 | 134.0 | 4.47% | 49.0 | 2.18% | motif file (matrix) |
pdf |
| 41 |  | ETS(ETS)/Promoter/Homer | 1e-5 | -1.248e+01 | 0.0000 | 40.0 | 1.33% | 5.7 | 0.26% | motif file (matrix) |
pdf |
| 42 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-5 | -1.191e+01 | 0.0001 | 109.0 | 3.63% | 37.1 | 1.65% | motif file (matrix) |
pdf |
| 43 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.170e+01 | 0.0001 | 74.0 | 2.47% | 20.9 | 0.93% | motif file (matrix) |
pdf |
| 44 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-4 | -1.086e+01 | 0.0002 | 122.0 | 4.07% | 46.0 | 2.05% | motif file (matrix) |
pdf |
| 45 |  | Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-4 | -1.086e+01 | 0.0002 | 386.0 | 12.87% | 207.6 | 9.25% | motif file (matrix) |
pdf |
| 46 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.063e+01 | 0.0002 | 79.0 | 2.63% | 25.0 | 1.11% | motif file (matrix) |
pdf |
| 47 | 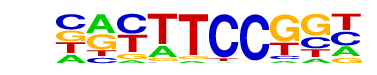 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-4 | -1.062e+01 | 0.0002 | 155.0 | 5.17% | 65.4 | 2.91% | motif file (matrix) |
pdf |
| 48 |  | Oct4:Sox17(POU,Homeobox,HMG)/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-4 | -1.040e+01 | 0.0002 | 38.0 | 1.27% | 6.7 | 0.30% | motif file (matrix) |
pdf |
| 49 |  | Tcf21(bHLH)/ArterySmoothMuscle-Tcf21-ChIP-Seq(GSE61369)/Homer | 1e-4 | -9.970e+00 | 0.0004 | 159.0 | 5.30% | 69.8 | 3.11% | motif file (matrix) |
pdf |
| 50 |  | HEB(bHLH)/mES-Heb-ChIP-Seq(GSE53233)/Homer | 1e-4 | -9.879e+00 | 0.0004 | 341.0 | 11.37% | 182.8 | 8.14% | motif file (matrix) |
pdf |
| 51 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -9.112e+00 | 0.0008 | 68.0 | 2.27% | 21.9 | 0.98% | motif file (matrix) |
pdf |
| 52 |  | MyoG(bHLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-3 | -9.015e+00 | 0.0009 | 178.0 | 5.93% | 83.4 | 3.71% | motif file (matrix) |
pdf |
| 53 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-3 | -8.830e+00 | 0.0011 | 29.0 | 0.97% | 4.1 | 0.18% | motif file (matrix) |
pdf |
| 54 |  | SeqBias: CG-repeat | 1e-3 | -8.805e+00 | 0.0011 | 375.0 | 12.50% | 209.1 | 9.31% | motif file (matrix) |
pdf |
| 55 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-3 | -8.315e+00 | 0.0017 | 391.0 | 13.03% | 222.4 | 9.90% | motif file (matrix) |
pdf |
| 56 |  | Pit1(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer | 1e-3 | -8.292e+00 | 0.0017 | 115.0 | 3.83% | 48.9 | 2.18% | motif file (matrix) |
pdf |
| 57 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-3 | -8.277e+00 | 0.0017 | 22.0 | 0.73% | 2.1 | 0.09% | motif file (matrix) |
pdf |
| 58 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-3 | -8.221e+00 | 0.0018 | 67.0 | 2.23% | 22.6 | 1.01% | motif file (matrix) |
pdf |
| 59 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-3 | -8.198e+00 | 0.0018 | 104.0 | 3.47% | 42.3 | 1.88% | motif file (matrix) |
pdf |
| 60 |  | E2A(bHLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-3 | -7.931e+00 | 0.0023 | 277.0 | 9.23% | 149.3 | 6.65% | motif file (matrix) |
pdf |
| 61 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-3 | -7.913e+00 | 0.0023 | 139.0 | 4.63% | 63.1 | 2.81% | motif file (matrix) |
pdf |
| 62 | 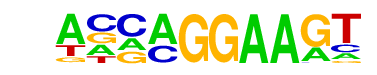 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-3 | -7.709e+00 | 0.0028 | 153.0 | 5.10% | 72.5 | 3.23% | motif file (matrix) |
pdf |
| 63 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-3 | -7.480e+00 | 0.0035 | 300.0 | 10.00% | 167.0 | 7.44% | motif file (matrix) |
pdf |
| 64 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-3 | -7.463e+00 | 0.0035 | 83.0 | 2.77% | 32.4 | 1.44% | motif file (matrix) |
pdf |
| 65 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-3 | -7.225e+00 | 0.0043 | 91.0 | 3.03% | 38.0 | 1.69% | motif file (matrix) |
pdf |
| 66 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -7.018e+00 | 0.0053 | 83.0 | 2.77% | 33.4 | 1.49% | motif file (matrix) |
pdf |
| 67 |  | ZBTB18(Zf)/HEK293-ZBTB18.GFP-ChIP-Seq(GSE58341)/Homer | 1e-3 | -6.991e+00 | 0.0053 | 90.0 | 3.00% | 37.2 | 1.65% | motif file (matrix) |
pdf |
| 68 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -6.893e+00 | 0.0058 | 16.0 | 0.53% | 1.0 | 0.05% | motif file (matrix) |
pdf |
| 69 |  | SeqBias: polyA-repeat | 1e-2 | -6.292e+00 | 0.0104 | 2640.0 | 88.00% | 1913.5 | 85.21% | motif file (matrix) |
pdf |
| 70 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-2 | -6.189e+00 | 0.0113 | 384.0 | 12.80% | 229.4 | 10.22% | motif file (matrix) |
pdf |
| 71 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-2 | -6.143e+00 | 0.0117 | 490.0 | 16.33% | 302.3 | 13.46% | motif file (matrix) |
pdf |
| 72 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-2 | -6.068e+00 | 0.0124 | 38.0 | 1.27% | 11.5 | 0.51% | motif file (matrix) |
pdf |
| 73 |  | DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer | 1e-2 | -6.014e+00 | 0.0130 | 134.0 | 4.47% | 66.0 | 2.94% | motif file (matrix) |
pdf |
| 74 | 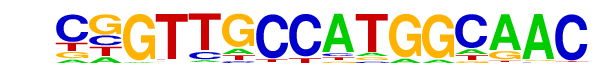 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -5.593e+00 | 0.0192 | 10.0 | 0.33% | 0.5 | 0.02% | motif file (matrix) |
pdf |
| 75 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-2 | -5.593e+00 | 0.0192 | 10.0 | 0.33% | 0.5 | 0.02% | motif file (matrix) |
pdf |
| 76 |  | SeqBias: G/A bias | 1e-2 | -5.574e+00 | 0.0193 | 2938.0 | 97.93% | 2171.3 | 96.69% | motif file (matrix) |
pdf |
| 77 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.541e+00 | 0.0197 | 152.0 | 5.07% | 79.8 | 3.55% | motif file (matrix) |
pdf |
| 78 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.517e+00 | 0.0199 | 46.0 | 1.53% | 16.2 | 0.72% | motif file (matrix) |
pdf |
| 79 |  | ETS:E-box(ETS,bHLH)/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-2 | -5.390e+00 | 0.0223 | 13.0 | 0.43% | 1.7 | 0.08% | motif file (matrix) |
pdf |
| 80 |  | Otx2(Homeobox)/EpiLC-Otx2-ChIP-Seq(GSE56098)/Homer | 1e-2 | -5.288e+00 | 0.0244 | 125.0 | 4.17% | 63.2 | 2.81% | motif file (matrix) |
pdf |
| 81 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.219e+00 | 0.0259 | 70.0 | 2.33% | 31.0 | 1.38% | motif file (matrix) |
pdf |
| 82 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-2 | -5.189e+00 | 0.0263 | 54.0 | 1.80% | 21.6 | 0.96% | motif file (matrix) |
pdf |
| 83 |  | Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.141e+00 | 0.0273 | 29.0 | 0.97% | 8.9 | 0.40% | motif file (matrix) |
pdf |
| 84 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-2 | -5.122e+00 | 0.0275 | 116.0 | 3.87% | 58.4 | 2.60% | motif file (matrix) |
pdf |
| 85 |  | Ascl1(bHLH)/NeuralTubes-Ascl1-ChIP-Seq(GSE55840)/Homer | 1e-2 | -4.703e+00 | 0.0413 | 263.0 | 8.77% | 156.2 | 6.96% | motif file (matrix) |
pdf |
| 86 |  | Ptf1a(bHLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-2 | -4.689e+00 | 0.0414 | 436.0 | 14.53% | 275.8 | 12.28% | motif file (matrix) |
pdf |
| 87 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.622e+00 | 0.0437 | 74.0 | 2.47% | 34.0 | 1.51% | motif file (matrix) |
pdf |













































































































































































