| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-8 | -1.881e+01 | 0.0000 | 89.0 | 5.71% | 28.8 | 1.89% | motif file (matrix) |
pdf |
| 2 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-7 | -1.764e+01 | 0.0000 | 285.0 | 18.29% | 172.2 | 11.26% | motif file (matrix) |
pdf |
| 3 |  | SeqBias: TA-repeat | 1e-7 | -1.668e+01 | 0.0000 | 508.0 | 32.61% | 366.5 | 23.97% | motif file (matrix) |
pdf |
| 4 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529)/Homer | 1e-4 | -1.089e+01 | 0.0018 | 74.0 | 4.75% | 31.2 | 2.04% | motif file (matrix) |
pdf |
| 5 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-4 | -1.040e+01 | 0.0024 | 127.0 | 8.15% | 70.2 | 4.59% | motif file (matrix) |
pdf |
| 6 | 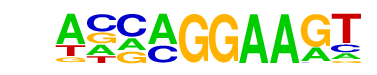 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-4 | -1.018e+01 | 0.0024 | 88.0 | 5.65% | 42.8 | 2.80% | motif file (matrix) |
pdf |
| 7 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-4 | -9.402e+00 | 0.0046 | 149.0 | 9.56% | 90.5 | 5.92% | motif file (matrix) |
pdf |
| 8 |  | Mef2b(MADS)/HEK293-Mef2b.V5-ChIP-Seq(GSE67450)/Homer | 1e-4 | -9.393e+00 | 0.0046 | 102.0 | 6.55% | 54.7 | 3.58% | motif file (matrix) |
pdf |
| 9 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-3 | -8.910e+00 | 0.0058 | 13.0 | 0.83% | 0.7 | 0.05% | motif file (matrix) |
pdf |
| 10 |  | Unknown2/Drosophila-Promoters/Homer | 1e-3 | -8.567e+00 | 0.0074 | 80.0 | 5.13% | 40.4 | 2.65% | motif file (matrix) |
pdf |
| 11 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-3 | -8.449e+00 | 0.0075 | 126.0 | 8.09% | 75.9 | 4.96% | motif file (matrix) |
pdf |
| 12 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.399e+00 | 0.0075 | 34.0 | 2.18% | 10.9 | 0.71% | motif file (matrix) |
pdf |
| 13 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-3 | -7.911e+00 | 0.0109 | 90.0 | 5.78% | 49.9 | 3.26% | motif file (matrix) |
pdf |
| 14 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-3 | -7.741e+00 | 0.0120 | 296.0 | 19.00% | 221.2 | 14.47% | motif file (matrix) |
pdf |
| 15 | 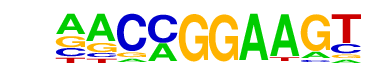 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-3 | -7.273e+00 | 0.0179 | 66.0 | 4.24% | 33.4 | 2.18% | motif file (matrix) |
pdf |
| 16 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-3 | -7.210e+00 | 0.0179 | 53.0 | 3.40% | 24.1 | 1.58% | motif file (matrix) |
pdf |
| 17 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-3 | -7.146e+00 | 0.0179 | 173.0 | 11.10% | 118.5 | 7.75% | motif file (matrix) |
pdf |
| 18 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-3 | -7.098e+00 | 0.0179 | 114.0 | 7.32% | 70.9 | 4.63% | motif file (matrix) |
pdf |
| 19 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.471e+00 | 0.0315 | 55.0 | 3.53% | 27.5 | 1.80% | motif file (matrix) |
pdf |
| 20 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-2 | -6.384e+00 | 0.0327 | 63.0 | 4.04% | 33.7 | 2.20% | motif file (matrix) |
pdf |
| 21 |  | Unknown6/Drosophila-Promoters/Homer | 1e-2 | -6.360e+00 | 0.0327 | 112.0 | 7.19% | 71.8 | 4.70% | motif file (matrix) |
pdf |
| 22 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.273e+00 | 0.0332 | 24.0 | 1.54% | 7.6 | 0.50% | motif file (matrix) |
pdf |
| 23 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-2 | -6.170e+00 | 0.0352 | 138.0 | 8.86% | 93.9 | 6.14% | motif file (matrix) |
pdf |
| 24 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.163e+00 | 0.0352 | 54.0 | 3.47% | 27.3 | 1.79% | motif file (matrix) |
pdf |
| 25 |  | ZNF165(Zf)/WHIM12-ZNF165-ChIP-Seq(GSE65937)/Homer | 1e-2 | -6.163e+00 | 0.0352 | 9.0 | 0.58% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 26 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.826e+00 | 0.0439 | 23.0 | 1.48% | 7.8 | 0.51% | motif file (matrix) |
pdf |
| 27 |  | Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer | 1e-2 | -5.708e+00 | 0.0476 | 185.0 | 11.87% | 135.6 | 8.87% | motif file (matrix) |
pdf |
| 28 | 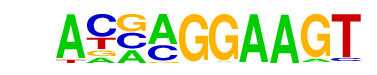 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-2 | -5.675e+00 | 0.0476 | 51.0 | 3.27% | 26.1 | 1.71% | motif file (matrix) |
pdf |
| 29 |  | SeqBias: CG bias | 1e-2 | -5.424e+00 | 0.0589 | 628.0 | 40.31% | 545.7 | 35.70% | motif file (matrix) |
pdf |
| 30 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -5.388e+00 | 0.0589 | 22.0 | 1.41% | 7.9 | 0.52% | motif file (matrix) |
pdf |
| 31 | 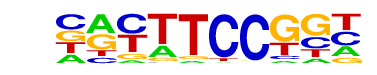 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.089e+00 | 0.0769 | 49.0 | 3.15% | 26.2 | 1.72% | motif file (matrix) |
pdf |
| 32 |  | ETS(ETS)/Promoter/Homer | 1e-2 | -4.965e+00 | 0.0844 | 14.0 | 0.90% | 3.9 | 0.25% | motif file (matrix) |
pdf |
| 33 |  | ZNF136(Zf)/HEK293-ZNF136.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -4.956e+00 | 0.0844 | 12.0 | 0.77% | 2.3 | 0.15% | motif file (matrix) |
pdf |
| 34 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -4.928e+00 | 0.0844 | 43.0 | 2.76% | 22.8 | 1.49% | motif file (matrix) |
pdf |
| 35 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-2 | -4.924e+00 | 0.0844 | 36.0 | 2.31% | 17.3 | 1.13% | motif file (matrix) |
pdf |
| 36 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-2 | -4.903e+00 | 0.0844 | 73.0 | 4.69% | 45.9 | 3.00% | motif file (matrix) |
pdf |
| 37 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-2 | -4.637e+00 | 0.1013 | 63.0 | 4.04% | 38.4 | 2.51% | motif file (matrix) |
pdf |









































































