| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-4 | -9.666e+00 | 0.0245 | 35.0 | 2.15% | 10.7 | 0.63% | motif file (matrix) |
pdf |
| 2 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-3 | -8.050e+00 | 0.0617 | 87.0 | 5.35% | 50.3 | 2.96% | motif file (matrix) |
pdf |
| 3 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.702e+00 | 0.0617 | 24.0 | 1.48% | 6.5 | 0.38% | motif file (matrix) |
pdf |
| 4 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.542e+00 | 0.0617 | 47.0 | 2.89% | 21.8 | 1.28% | motif file (matrix) |
pdf |
| 5 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.480e+00 | 0.1187 | 18.0 | 1.11% | 4.9 | 0.29% | motif file (matrix) |
pdf |
| 6 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.112e+00 | 0.1430 | 43.0 | 2.64% | 21.7 | 1.28% | motif file (matrix) |
pdf |
| 7 |  | FOXA1:AR(Forkhead,NR)/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.867e+00 | 0.1565 | 13.0 | 0.80% | 2.7 | 0.16% | motif file (matrix) |
pdf |
| 8 |  | E-box/Arabidopsis-Promoters/Homer | 1e-2 | -5.719e+00 | 0.1587 | 20.0 | 1.23% | 6.2 | 0.36% | motif file (matrix) |
pdf |
| 9 |  | NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.645e+00 | 0.1587 | 72.0 | 4.43% | 45.3 | 2.67% | motif file (matrix) |
pdf |
| 10 | 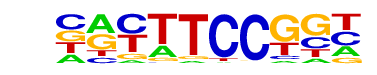 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.617e+00 | 0.1587 | 52.0 | 3.20% | 29.7 | 1.75% | motif file (matrix) |
pdf |
| 11 | 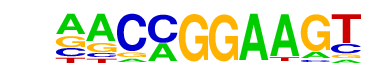 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -5.487e+00 | 0.1587 | 75.0 | 4.61% | 48.0 | 2.83% | motif file (matrix) |
pdf |
| 12 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-2 | -5.282e+00 | 0.1640 | 12.0 | 0.74% | 2.2 | 0.13% | motif file (matrix) |
pdf |
| 13 |  | SUT1?/SacCer-Promoters/Homer | 1e-2 | -5.253e+00 | 0.1640 | 299.0 | 18.38% | 255.7 | 15.05% | motif file (matrix) |
pdf |
| 14 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.139e+00 | 0.1640 | 44.0 | 2.70% | 24.4 | 1.44% | motif file (matrix) |
pdf |
| 15 |  | PAX6(Paired,Homeobox)/Forebrain-Pax6-ChIP-Seq(GSE66961)/Homer | 1e-2 | -5.012e+00 | 0.1718 | 7.0 | 0.43% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 16 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-2 | -4.795e+00 | 0.2001 | 78.0 | 4.79% | 53.9 | 3.17% | motif file (matrix) |
pdf |
| 17 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.778e+00 | 0.2001 | 31.0 | 1.91% | 15.2 | 0.89% | motif file (matrix) |
pdf |

































