| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-5 | -1.247e+01 | 0.0015 | 82.0 | 9.12% | 32.1 | 3.80% | motif file (matrix) |
pdf |
| 2 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-4 | -9.273e+00 | 0.0182 | 138.0 | 15.35% | 79.2 | 9.38% | motif file (matrix) |
pdf |
| 3 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-3 | -9.162e+00 | 0.0182 | 29.0 | 3.23% | 6.1 | 0.72% | motif file (matrix) |
pdf |
| 4 |  | SeqBias: TA-repeat | 1e-3 | -7.778e+00 | 0.0405 | 229.0 | 25.47% | 158.2 | 18.73% | motif file (matrix) |
pdf |
| 5 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-3 | -7.477e+00 | 0.0438 | 57.0 | 6.34% | 25.8 | 3.06% | motif file (matrix) |
pdf |
| 6 |  | Srebp2(bHLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.260e+00 | 0.0453 | 14.0 | 1.56% | 1.0 | 0.12% | motif file (matrix) |
pdf |
| 7 |  | Unknown2/Drosophila-Promoters/Homer | 1e-3 | -6.983e+00 | 0.0513 | 54.0 | 6.01% | 24.7 | 2.92% | motif file (matrix) |
pdf |
| 8 |  | Sox15(HMG)/CPA-Sox15-ChIP-Seq(GSE62909)/Homer | 1e-2 | -6.660e+00 | 0.0620 | 47.0 | 5.23% | 20.7 | 2.45% | motif file (matrix) |
pdf |
| 9 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-2 | -6.152e+00 | 0.0916 | 67.0 | 7.45% | 35.4 | 4.19% | motif file (matrix) |
pdf |
| 10 |  | HEB(bHLH)/mES-Heb-ChIP-Seq(GSE53233)/Homer | 1e-2 | -6.150e+00 | 0.0916 | 60.0 | 6.67% | 30.4 | 3.60% | motif file (matrix) |
pdf |
| 11 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-2 | -6.143e+00 | 0.0916 | 33.0 | 3.67% | 12.2 | 1.44% | motif file (matrix) |
pdf |
| 12 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.056e+00 | 0.0916 | 12.0 | 1.33% | 1.5 | 0.18% | motif file (matrix) |
pdf |
| 13 | 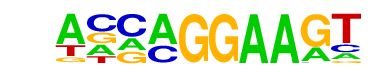 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.006e+00 | 0.0916 | 48.0 | 5.34% | 22.4 | 2.65% | motif file (matrix) |
pdf |
| 14 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-2 | -5.796e+00 | 0.0916 | 150.0 | 16.69% | 101.5 | 12.01% | motif file (matrix) |
pdf |
| 15 |  | SeqBias: GA-repeat | 1e-2 | -5.743e+00 | 0.0916 | 300.0 | 33.37% | 230.7 | 27.30% | motif file (matrix) |
pdf |
| 16 |  | DMRT1(DM)/Testis-DMRT1-ChIP-Seq(GSE64892)/Homer | 1e-2 | -5.282e+00 | 0.1092 | 13.0 | 1.45% | 2.9 | 0.34% | motif file (matrix) |
pdf |
| 17 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -5.282e+00 | 0.1092 | 13.0 | 1.45% | 2.7 | 0.32% | motif file (matrix) |
pdf |
| 18 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-2 | -5.282e+00 | 0.1092 | 13.0 | 1.45% | 2.2 | 0.26% | motif file (matrix) |
pdf |
| 19 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-2 | -5.241e+00 | 0.1092 | 54.0 | 6.01% | 28.5 | 3.38% | motif file (matrix) |
pdf |
| 20 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.191e+00 | 0.1092 | 32.0 | 3.56% | 13.2 | 1.56% | motif file (matrix) |
pdf |
| 21 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.168e+00 | 0.1092 | 66.0 | 7.34% | 37.9 | 4.48% | motif file (matrix) |
pdf |
| 22 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-2 | -5.112e+00 | 0.1092 | 112.0 | 12.46% | 73.4 | 8.68% | motif file (matrix) |
pdf |
| 23 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.105e+00 | 0.1092 | 59.0 | 6.56% | 32.9 | 3.90% | motif file (matrix) |
pdf |
| 24 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-2 | -4.959e+00 | 0.1132 | 46.0 | 5.12% | 23.8 | 2.82% | motif file (matrix) |
pdf |
| 25 |  | Tcf21(bHLH)/ArterySmoothMuscle-Tcf21-ChIP-Seq(GSE61369)/Homer | 1e-2 | -4.862e+00 | 0.1197 | 18.0 | 2.00% | 5.1 | 0.60% | motif file (matrix) |
pdf |
| 26 |  | Ascl1(bHLH)/NeuralTubes-Ascl1-ChIP-Seq(GSE55840)/Homer | 1e-2 | -4.835e+00 | 0.1197 | 34.0 | 3.78% | 15.7 | 1.86% | motif file (matrix) |
pdf |
| 27 |  | Srebp1a(bHLH)/HepG2-Srebp1a-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.782e+00 | 0.1197 | 16.0 | 1.78% | 4.3 | 0.51% | motif file (matrix) |
pdf |
| 28 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-2 | -4.782e+00 | 0.1197 | 16.0 | 1.78% | 4.9 | 0.58% | motif file (matrix) |
pdf |
| 29 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-2 | -4.735e+00 | 0.1197 | 14.0 | 1.56% | 3.7 | 0.43% | motif file (matrix) |
pdf |
| 30 |  | HNF4a(NR),DR1/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-2 | -4.735e+00 | 0.1197 | 14.0 | 1.56% | 3.5 | 0.42% | motif file (matrix) |
pdf |
| 31 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-2 | -4.668e+00 | 0.1197 | 21.0 | 2.34% | 7.4 | 0.88% | motif file (matrix) |
pdf |





























































