| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-3 | -8.912e+00 | 0.0522 | 18.0 | 1.14% | 2.5 | 0.15% | motif file (matrix) |
pdf |
| 2 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-3 | -8.606e+00 | 0.0522 | 12.0 | 0.76% | 0.9 | 0.05% | motif file (matrix) |
pdf |
| 3 |  | Unknown6/Drosophila-Promoters/Homer | 1e-3 | -7.867e+00 | 0.0522 | 124.0 | 7.85% | 81.4 | 4.93% | motif file (matrix) |
pdf |
| 4 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-3 | -7.702e+00 | 0.0522 | 34.0 | 2.15% | 12.1 | 0.73% | motif file (matrix) |
pdf |
| 5 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-2 | -6.362e+00 | 0.1336 | 293.0 | 18.54% | 242.1 | 14.66% | motif file (matrix) |
pdf |
| 6 |  | E-box/Arabidopsis-Promoters/Homer | 1e-2 | -6.069e+00 | 0.1491 | 19.0 | 1.20% | 5.8 | 0.35% | motif file (matrix) |
pdf |
| 7 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.016e+00 | 0.1491 | 30.0 | 1.90% | 12.5 | 0.76% | motif file (matrix) |
pdf |
| 8 |  | Mouse_Recombination_Hotspot(Zf)/Testis-DMC1-ChIP-Seq(GSE24438)/Homer | 1e-2 | -5.732e+00 | 0.1568 | 8.0 | 0.51% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 9 | 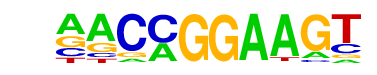 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -5.404e+00 | 0.1935 | 71.0 | 4.49% | 45.6 | 2.76% | motif file (matrix) |
pdf |
| 10 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.392e+00 | 0.1935 | 24.0 | 1.52% | 9.4 | 0.57% | motif file (matrix) |
pdf |
| 11 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.080e+00 | 0.2189 | 17.0 | 1.08% | 5.9 | 0.36% | motif file (matrix) |
pdf |





















