| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-4 | -1.138e+01 | 0.0044 | 116.0 | 13.43% | 55.1 | 7.02% | motif file (matrix) |
pdf |
| 2 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-4 | -1.128e+01 | 0.0044 | 84.0 | 9.72% | 34.9 | 4.44% | motif file (matrix) |
pdf |
| 3 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-4 | -9.754e+00 | 0.0075 | 15.0 | 1.74% | 0.5 | 0.07% | motif file (matrix) |
pdf |
| 4 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-3 | -8.243e+00 | 0.0255 | 116.0 | 13.43% | 63.8 | 8.11% | motif file (matrix) |
pdf |
| 5 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-3 | -7.793e+00 | 0.0319 | 12.0 | 1.39% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 6 | 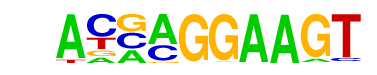 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-3 | -7.382e+00 | 0.0402 | 26.0 | 3.01% | 6.5 | 0.83% | motif file (matrix) |
pdf |
| 7 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-3 | -7.359e+00 | 0.0402 | 152.0 | 17.59% | 93.9 | 11.94% | motif file (matrix) |
pdf |
| 8 | 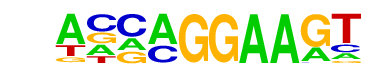 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.893e+00 | 0.0491 | 49.0 | 5.67% | 20.1 | 2.56% | motif file (matrix) |
pdf |
| 9 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-2 | -5.989e+00 | 0.1077 | 21.0 | 2.43% | 5.4 | 0.69% | motif file (matrix) |
pdf |
| 10 |  | Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer | 1e-2 | -5.766e+00 | 0.1213 | 125.0 | 14.47% | 78.9 | 10.03% | motif file (matrix) |
pdf |
| 11 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -5.685e+00 | 0.1213 | 31.0 | 3.59% | 11.1 | 1.42% | motif file (matrix) |
pdf |
| 12 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-2 | -5.558e+00 | 0.1244 | 61.0 | 7.06% | 31.8 | 4.05% | motif file (matrix) |
pdf |
| 13 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.513e+00 | 0.1244 | 18.0 | 2.08% | 4.8 | 0.61% | motif file (matrix) |
pdf |
| 14 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.304e+00 | 0.1374 | 11.0 | 1.27% | 1.3 | 0.17% | motif file (matrix) |
pdf |
| 15 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-2 | -5.186e+00 | 0.1443 | 8.0 | 0.93% | 0.6 | 0.07% | motif file (matrix) |
pdf |
| 16 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.986e+00 | 0.1653 | 40.0 | 4.63% | 18.6 | 2.36% | motif file (matrix) |
pdf |
| 17 |  | SeqBias: TA-repeat | 1e-2 | -4.740e+00 | 0.1989 | 209.0 | 24.19% | 151.7 | 19.30% | motif file (matrix) |
pdf |
| 18 | 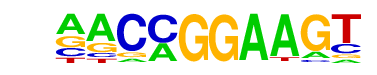 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -4.693e+00 | 0.1989 | 42.0 | 4.86% | 20.5 | 2.61% | motif file (matrix) |
pdf |



































