| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-95 | -2.202e+02 | 0.0000 | 811.0 | 27.03% | 122.3 | 5.69% | motif file (matrix) |
pdf |
| 2 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-83 | -1.916e+02 | 0.0000 | 706.0 | 23.53% | 103.2 | 4.80% | motif file (matrix) |
pdf |
| 3 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-77 | -1.777e+02 | 0.0000 | 652.0 | 21.73% | 93.9 | 4.37% | motif file (matrix) |
pdf |
| 4 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-67 | -1.551e+02 | 0.0000 | 475.0 | 15.83% | 47.9 | 2.23% | motif file (matrix) |
pdf |
| 5 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-46 | -1.070e+02 | 0.0000 | 383.0 | 12.77% | 50.0 | 2.32% | motif file (matrix) |
pdf |
| 6 |  | Sox15(HMG)/CPA-Sox15-ChIP-Seq(GSE62909)/Homer | 1e-43 | -1.002e+02 | 0.0000 | 454.0 | 15.13% | 81.7 | 3.80% | motif file (matrix) |
pdf |
| 7 |  | OCT4-SOX2-TCF-NANOG(POU,Homeobox,HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-30 | -7.077e+01 | 0.0000 | 188.0 | 6.27% | 12.9 | 0.60% | motif file (matrix) |
pdf |
| 8 |  | Zic(Zf)/Cerebellum-ZIC1.2-ChIP-Seq(GSE60731)/Homer | 1e-28 | -6.577e+01 | 0.0000 | 341.0 | 11.37% | 69.0 | 3.21% | motif file (matrix) |
pdf |
| 9 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-23 | -5.425e+01 | 0.0000 | 304.0 | 10.13% | 66.9 | 3.11% | motif file (matrix) |
pdf |
| 10 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-21 | -4.841e+01 | 0.0000 | 877.0 | 29.23% | 383.0 | 17.82% | motif file (matrix) |
pdf |
| 11 |  | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-19 | -4.573e+01 | 0.0000 | 186.0 | 6.20% | 28.9 | 1.34% | motif file (matrix) |
pdf |
| 12 |  | Sox9(HMG)/Limb-SOX9-ChIP-Seq(GSE73225)/Homer | 1e-18 | -4.155e+01 | 0.0000 | 260.0 | 8.67% | 62.2 | 2.90% | motif file (matrix) |
pdf |
| 13 |  | Oct6(POU,Homeobox)/NPC-Oct6-ChIP-Seq(GSE35496)/Homer | 1e-15 | -3.597e+01 | 0.0000 | 146.0 | 4.87% | 22.1 | 1.03% | motif file (matrix) |
pdf |
| 14 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-14 | -3.437e+01 | 0.0000 | 219.0 | 7.30% | 53.5 | 2.49% | motif file (matrix) |
pdf |
| 15 |  | DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer | 1e-14 | -3.265e+01 | 0.0000 | 179.0 | 5.97% | 38.3 | 1.78% | motif file (matrix) |
pdf |
| 16 |  | Brn1(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer | 1e-13 | -3.100e+01 | 0.0000 | 120.0 | 4.00% | 17.2 | 0.80% | motif file (matrix) |
pdf |
| 17 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-13 | -3.088e+01 | 0.0000 | 139.0 | 4.63% | 24.1 | 1.12% | motif file (matrix) |
pdf |
| 18 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-12 | -2.773e+01 | 0.0000 | 540.0 | 18.00% | 235.3 | 10.95% | motif file (matrix) |
pdf |
| 19 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-11 | -2.559e+01 | 0.0000 | 960.0 | 32.00% | 503.6 | 23.43% | motif file (matrix) |
pdf |
| 20 |  | TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer | 1e-11 | -2.541e+01 | 0.0000 | 130.0 | 4.33% | 26.9 | 1.25% | motif file (matrix) |
pdf |
| 21 |  | Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-10 | -2.500e+01 | 0.0000 | 100.0 | 3.33% | 15.4 | 0.72% | motif file (matrix) |
pdf |
| 22 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-10 | -2.468e+01 | 0.0000 | 102.0 | 3.40% | 16.5 | 0.77% | motif file (matrix) |
pdf |
| 23 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-9 | -2.137e+01 | 0.0000 | 179.0 | 5.97% | 53.3 | 2.48% | motif file (matrix) |
pdf |
| 24 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-8 | -2.030e+01 | 0.0000 | 351.0 | 11.70% | 146.7 | 6.83% | motif file (matrix) |
pdf |
| 25 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-8 | -1.962e+01 | 0.0000 | 50.0 | 1.67% | 3.9 | 0.18% | motif file (matrix) |
pdf |
| 26 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-8 | -1.926e+01 | 0.0000 | 441.0 | 14.70% | 201.2 | 9.36% | motif file (matrix) |
pdf |
| 27 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-7 | -1.831e+01 | 0.0000 | 152.0 | 5.07% | 45.2 | 2.10% | motif file (matrix) |
pdf |
| 28 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.822e+01 | 0.0000 | 204.0 | 6.80% | 71.8 | 3.34% | motif file (matrix) |
pdf |
| 29 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.807e+01 | 0.0000 | 147.0 | 4.90% | 43.9 | 2.04% | motif file (matrix) |
pdf |
| 30 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-7 | -1.800e+01 | 0.0000 | 335.0 | 11.17% | 143.9 | 6.70% | motif file (matrix) |
pdf |
| 31 |  | Lhx1(Homeobox)/EmbryoCarcinoma-Lhx1-ChIP-Seq(GSE70957)/Homer | 1e-7 | -1.738e+01 | 0.0000 | 246.0 | 8.20% | 95.3 | 4.44% | motif file (matrix) |
pdf |
| 32 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-7 | -1.737e+01 | 0.0000 | 86.0 | 2.87% | 17.1 | 0.79% | motif file (matrix) |
pdf |
| 33 | 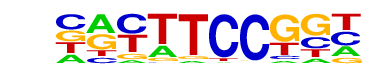 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-7 | -1.730e+01 | 0.0000 | 169.0 | 5.63% | 55.4 | 2.58% | motif file (matrix) |
pdf |
| 34 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-7 | -1.649e+01 | 0.0000 | 156.0 | 5.20% | 50.9 | 2.37% | motif file (matrix) |
pdf |
| 35 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-7 | -1.630e+01 | 0.0000 | 234.0 | 7.80% | 91.7 | 4.27% | motif file (matrix) |
pdf |
| 36 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-6 | -1.598e+01 | 0.0000 | 276.0 | 9.20% | 115.4 | 5.37% | motif file (matrix) |
pdf |
| 37 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.506e+01 | 0.0000 | 105.0 | 3.50% | 28.2 | 1.31% | motif file (matrix) |
pdf |
| 38 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-6 | -1.383e+01 | 0.0000 | 156.0 | 5.20% | 55.0 | 2.56% | motif file (matrix) |
pdf |
| 39 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-5 | -1.379e+01 | 0.0000 | 122.0 | 4.07% | 38.2 | 1.78% | motif file (matrix) |
pdf |
| 40 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-5 | -1.348e+01 | 0.0000 | 127.0 | 4.23% | 41.2 | 1.92% | motif file (matrix) |
pdf |
| 41 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-5 | -1.337e+01 | 0.0000 | 347.0 | 11.57% | 164.4 | 7.65% | motif file (matrix) |
pdf |
| 42 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-5 | -1.292e+01 | 0.0000 | 59.0 | 1.97% | 12.0 | 0.56% | motif file (matrix) |
pdf |
| 43 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-5 | -1.191e+01 | 0.0001 | 22.0 | 0.73% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 44 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-5 | -1.182e+01 | 0.0001 | 146.0 | 4.87% | 54.2 | 2.52% | motif file (matrix) |
pdf |
| 45 |  | SeqBias: CG bias | 1e-4 | -1.110e+01 | 0.0001 | 2014.0 | 67.13% | 1320.8 | 61.47% | motif file (matrix) |
pdf |
| 46 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-4 | -1.086e+01 | 0.0002 | 142.0 | 4.73% | 54.8 | 2.55% | motif file (matrix) |
pdf |
| 47 |  | Tcf21(bHLH)/ArterySmoothMuscle-Tcf21-ChIP-Seq(GSE61369)/Homer | 1e-4 | -1.083e+01 | 0.0002 | 164.0 | 5.47% | 66.1 | 3.08% | motif file (matrix) |
pdf |
| 48 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-4 | -1.027e+01 | 0.0003 | 132.0 | 4.40% | 50.1 | 2.33% | motif file (matrix) |
pdf |
| 49 |  | Oct4:Sox17(POU,Homeobox,HMG)/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-4 | -1.026e+01 | 0.0003 | 27.0 | 0.90% | 2.9 | 0.13% | motif file (matrix) |
pdf |
| 50 |  | Ascl1(bHLH)/NeuralTubes-Ascl1-ChIP-Seq(GSE55840)/Homer | 1e-4 | -1.005e+01 | 0.0003 | 244.0 | 8.13% | 114.2 | 5.31% | motif file (matrix) |
pdf |
| 51 | 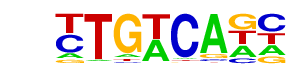 | Tgif1(Homeobox)/mES-Tgif1-ChIP-Seq(GSE55404)/Homer | 1e-4 | -9.951e+00 | 0.0004 | 391.0 | 13.03% | 204.9 | 9.53% | motif file (matrix) |
pdf |
| 52 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-4 | -9.694e+00 | 0.0005 | 83.0 | 2.77% | 26.4 | 1.23% | motif file (matrix) |
pdf |
| 53 |  | RBPJ:Ebox(?,bHLH)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-4 | -9.599e+00 | 0.0005 | 50.0 | 1.67% | 11.5 | 0.54% | motif file (matrix) |
pdf |
| 54 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-4 | -9.451e+00 | 0.0006 | 61.0 | 2.03% | 16.1 | 0.75% | motif file (matrix) |
pdf |
| 55 |  | SeqBias: GA-repeat | 1e-4 | -9.367e+00 | 0.0006 | 1134.0 | 37.80% | 702.4 | 32.69% | motif file (matrix) |
pdf |
| 56 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-4 | -9.317e+00 | 0.0006 | 211.0 | 7.03% | 97.9 | 4.56% | motif file (matrix) |
pdf |
| 57 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-4 | -9.223e+00 | 0.0007 | 28.0 | 0.93% | 3.0 | 0.14% | motif file (matrix) |
pdf |
| 58 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-3 | -9.170e+00 | 0.0007 | 97.0 | 3.23% | 34.4 | 1.60% | motif file (matrix) |
pdf |
| 59 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-3 | -9.170e+00 | 0.0007 | 75.0 | 2.50% | 23.6 | 1.10% | motif file (matrix) |
pdf |
| 60 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-3 | -8.762e+00 | 0.0010 | 61.0 | 2.03% | 17.3 | 0.80% | motif file (matrix) |
pdf |
| 61 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-3 | -8.703e+00 | 0.0011 | 99.0 | 3.30% | 36.2 | 1.69% | motif file (matrix) |
pdf |
| 62 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-3 | -8.630e+00 | 0.0011 | 69.0 | 2.30% | 21.5 | 1.00% | motif file (matrix) |
pdf |
| 63 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.583e+00 | 0.0012 | 81.0 | 2.70% | 27.2 | 1.27% | motif file (matrix) |
pdf |
| 64 |  | Tgif2(Homeobox)/mES-Tgif2-ChIP-Seq(GSE55404)/Homer | 1e-3 | -8.482e+00 | 0.0013 | 393.0 | 13.10% | 212.9 | 9.91% | motif file (matrix) |
pdf |
| 65 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.360e+00 | 0.0014 | 66.0 | 2.20% | 20.5 | 0.95% | motif file (matrix) |
pdf |
| 66 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-3 | -8.324e+00 | 0.0014 | 202.0 | 6.73% | 95.2 | 4.43% | motif file (matrix) |
pdf |
| 67 |  | ETS(ETS)/Promoter/Homer | 1e-3 | -8.206e+00 | 0.0016 | 46.0 | 1.53% | 11.6 | 0.54% | motif file (matrix) |
pdf |
| 68 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-3 | -7.933e+00 | 0.0020 | 118.0 | 3.93% | 48.6 | 2.26% | motif file (matrix) |
pdf |
| 69 |  | SeqBias: polyC-repeat | 1e-3 | -7.880e+00 | 0.0021 | 2824.0 | 94.13% | 1969.3 | 91.65% | motif file (matrix) |
pdf |
| 70 |  | PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer | 1e-3 | -7.867e+00 | 0.0021 | 45.0 | 1.50% | 12.0 | 0.56% | motif file (matrix) |
pdf |
| 71 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.767e+00 | 0.0023 | 68.0 | 2.27% | 22.4 | 1.04% | motif file (matrix) |
pdf |
| 72 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-3 | -7.733e+00 | 0.0024 | 142.0 | 4.73% | 62.7 | 2.92% | motif file (matrix) |
pdf |
| 73 | 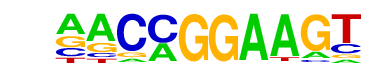 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-3 | -7.639e+00 | 0.0026 | 203.0 | 6.77% | 99.0 | 4.61% | motif file (matrix) |
pdf |
| 74 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.605e+00 | 0.0026 | 383.0 | 12.77% | 210.5 | 9.80% | motif file (matrix) |
pdf |
| 75 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-3 | -7.603e+00 | 0.0026 | 249.0 | 8.30% | 126.8 | 5.90% | motif file (matrix) |
pdf |
| 76 |  | E2A(bHLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-3 | -7.588e+00 | 0.0026 | 244.0 | 8.13% | 123.7 | 5.76% | motif file (matrix) |
pdf |
| 77 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-3 | -7.458e+00 | 0.0029 | 32.0 | 1.07% | 6.3 | 0.29% | motif file (matrix) |
pdf |
| 78 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-2 | -6.887e+00 | 0.0051 | 28.0 | 0.93% | 5.2 | 0.24% | motif file (matrix) |
pdf |
| 79 |  | SeqBias: polyA-repeat | 1e-2 | -6.871e+00 | 0.0051 | 2526.0 | 84.20% | 1737.7 | 80.87% | motif file (matrix) |
pdf |
| 80 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-2 | -6.774e+00 | 0.0055 | 74.0 | 2.47% | 27.5 | 1.28% | motif file (matrix) |
pdf |
| 81 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-2 | -6.413e+00 | 0.0078 | 61.0 | 2.03% | 21.3 | 0.99% | motif file (matrix) |
pdf |
| 82 |  | SUT1?/SacCer-Promoters/Homer | 1e-2 | -6.279e+00 | 0.0088 | 1459.0 | 48.63% | 956.0 | 44.49% | motif file (matrix) |
pdf |
| 83 |  | Tcf12(bHLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.031e+00 | 0.0112 | 178.0 | 5.93% | 89.4 | 4.16% | motif file (matrix) |
pdf |
| 84 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-2 | -5.964e+00 | 0.0118 | 28.0 | 0.93% | 7.0 | 0.32% | motif file (matrix) |
pdf |
| 85 |  | SeqBias: CG-repeat | 1e-2 | -5.511e+00 | 0.0184 | 622.0 | 20.73% | 381.6 | 17.76% | motif file (matrix) |
pdf |
| 86 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.500e+00 | 0.0184 | 92.0 | 3.07% | 40.1 | 1.87% | motif file (matrix) |
pdf |
| 87 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.382e+00 | 0.0205 | 79.0 | 2.63% | 33.2 | 1.54% | motif file (matrix) |
pdf |
| 88 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.283e+00 | 0.0223 | 43.0 | 1.43% | 14.8 | 0.69% | motif file (matrix) |
pdf |
| 89 |  | Ap4(bHLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-2 | -5.254e+00 | 0.0227 | 205.0 | 6.83% | 109.5 | 5.10% | motif file (matrix) |
pdf |
| 90 |  | HLH-1(bHLH)/cElegans-Embryo-HLH1-ChIP-Seq(modEncode)/Homer | 1e-2 | -5.215e+00 | 0.0234 | 118.0 | 3.93% | 56.2 | 2.61% | motif file (matrix) |
pdf |
| 91 |  | Ptf1a(bHLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-2 | -5.163e+00 | 0.0243 | 392.0 | 13.07% | 231.0 | 10.75% | motif file (matrix) |
pdf |
| 92 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-2 | -5.109e+00 | 0.0254 | 114.0 | 3.80% | 54.8 | 2.55% | motif file (matrix) |
pdf |
| 93 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -5.050e+00 | 0.0267 | 63.0 | 2.10% | 25.4 | 1.18% | motif file (matrix) |
pdf |
| 94 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-2 | -4.866e+00 | 0.0317 | 64.0 | 2.13% | 26.1 | 1.21% | motif file (matrix) |
pdf |
| 95 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.865e+00 | 0.0317 | 9.0 | 0.30% | 0.4 | 0.02% | motif file (matrix) |
pdf |
| 96 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-2 | -4.789e+00 | 0.0335 | 292.0 | 9.73% | 167.2 | 7.78% | motif file (matrix) |
pdf |
| 97 |  | Unknown2/Drosophila-Promoters/Homer | 1e-2 | -4.653e+00 | 0.0380 | 154.0 | 5.13% | 80.1 | 3.73% | motif file (matrix) |
pdf |

































































































































































































